Great Apes Chimp
Report card for Correlation and Tree Mapping on Chimp neocortex (Jorstad et al. 2023)
Overview
A taxonomy was initially built using the chimp neocortex single nucleus dataset. In building the taxonomy, 1000 binary marker genes were selected based on their gene expression from the single-cell transcriptome. Subsequently, the dataset was mapped to itself, termed self-projection, for evaluating the ideal performances of correlation and tree mapping algorithms.
Quantitative analysis
The analysis evaluates the predictions of correlation and tree mappings in determining cluster labels in a self-projection evaluation.
| Annotaion | F1-score |
|---|---|
| Cluster Correlation Mapping | 0.890 |
| Cluster Tree Mapping | 0.868 |
Correlation Mapping
-
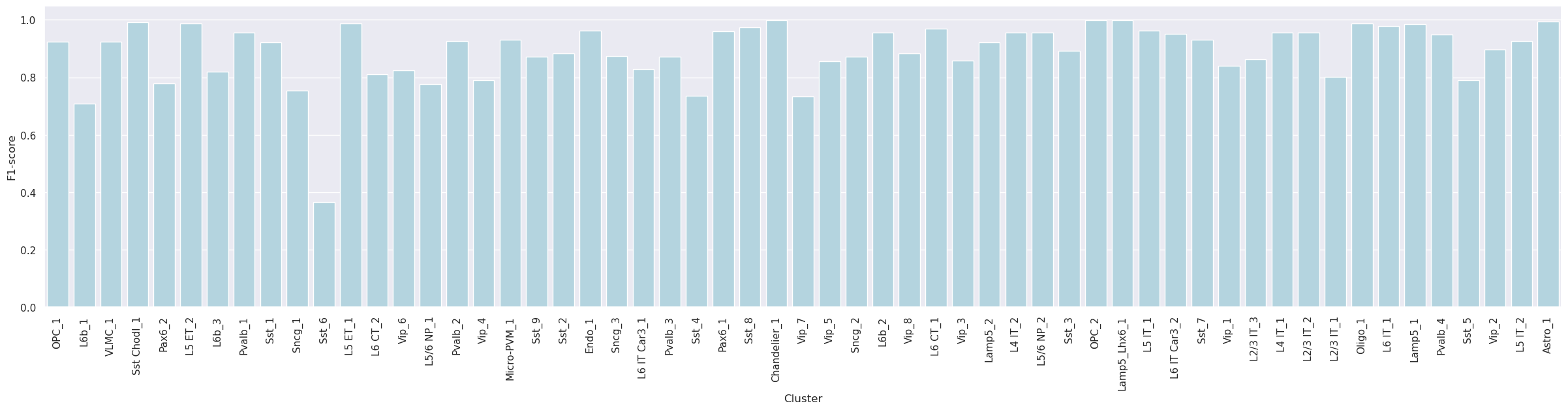
Label-wise F1-score
-
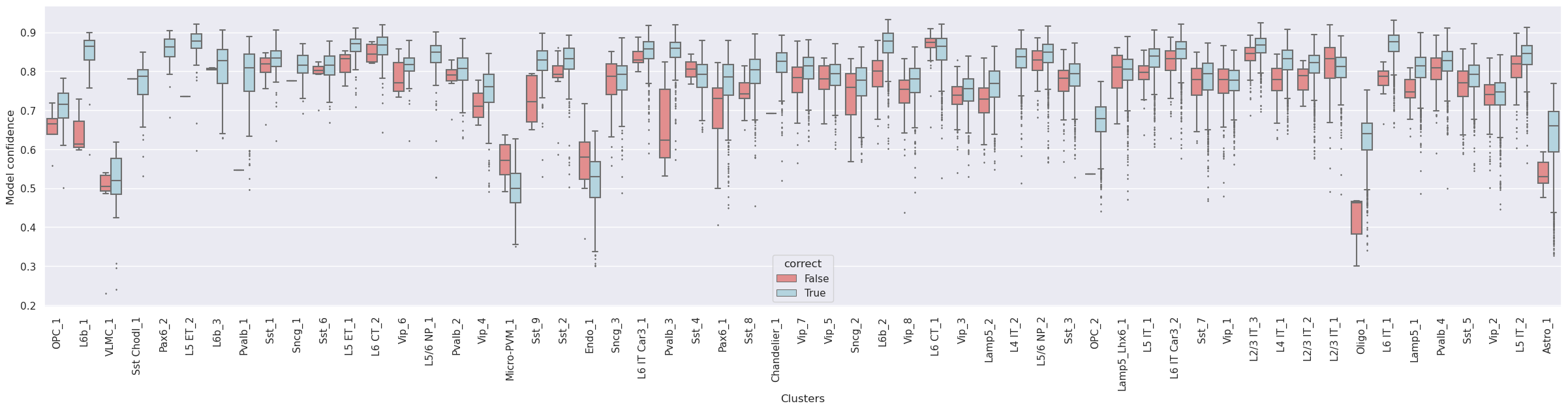
Confidence values for correctly and incorrectly assigned labels
-
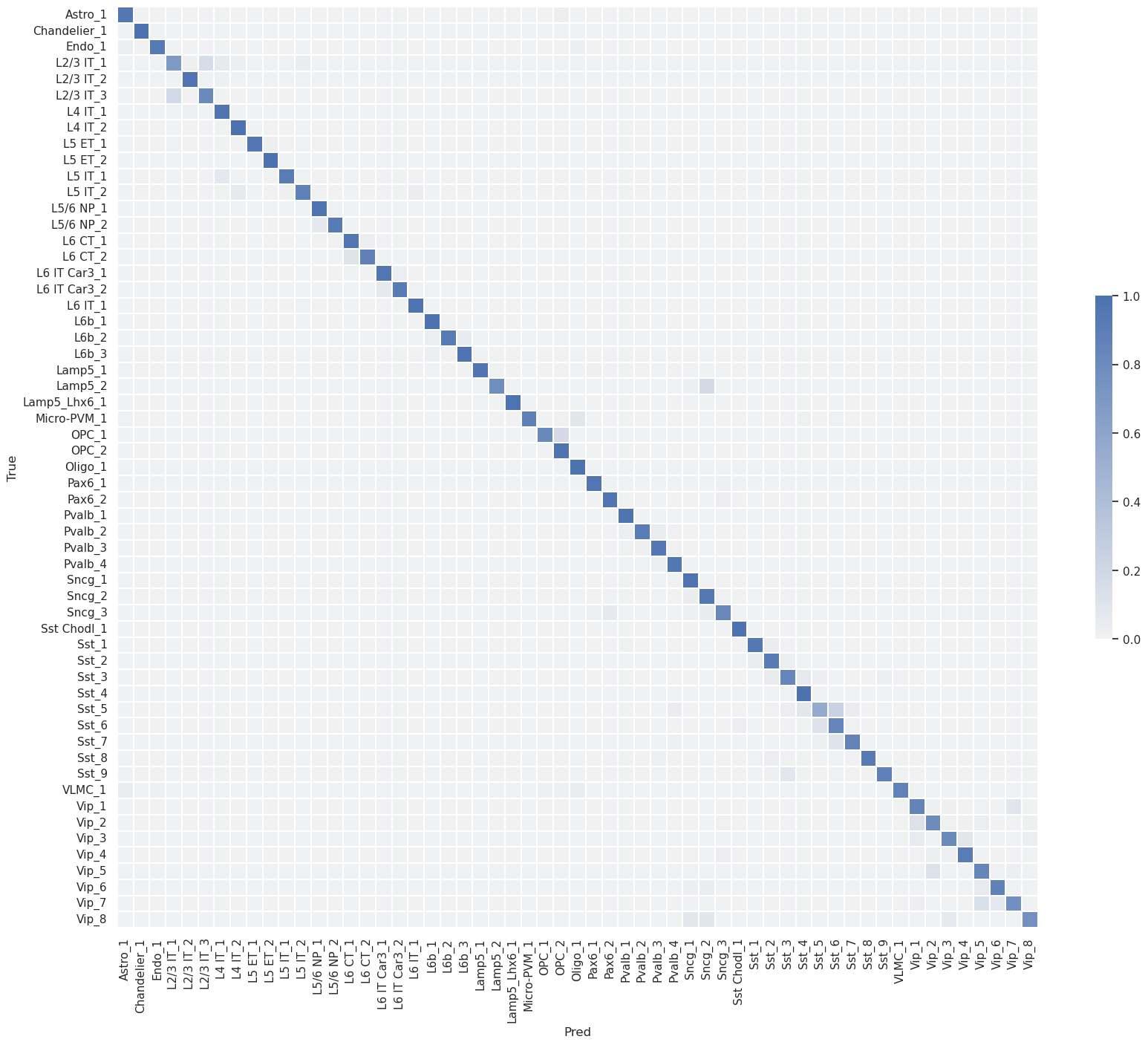
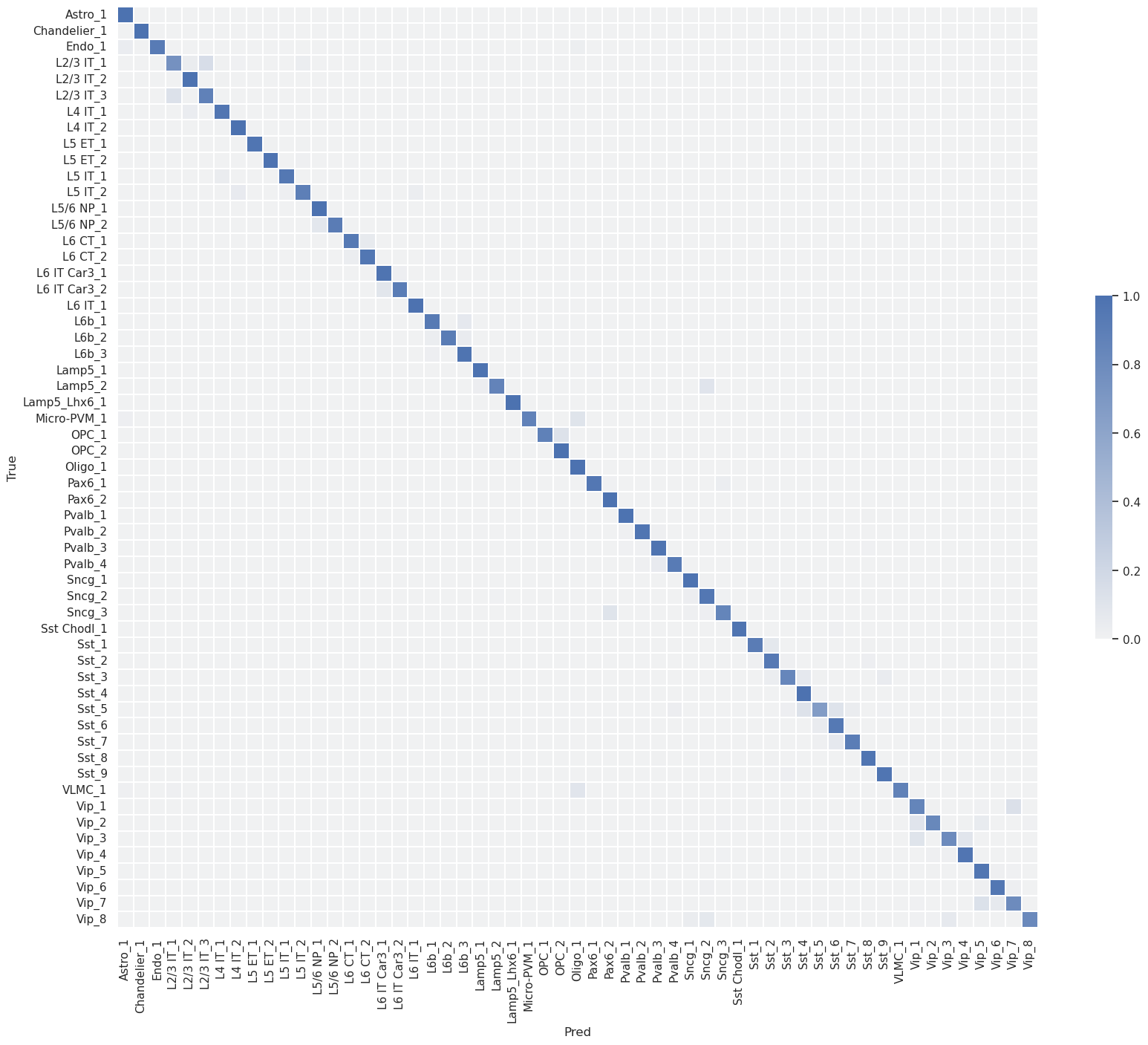
Confusion matrix (row-normalized)
Tree Mapping
-
Label-wise F1-score
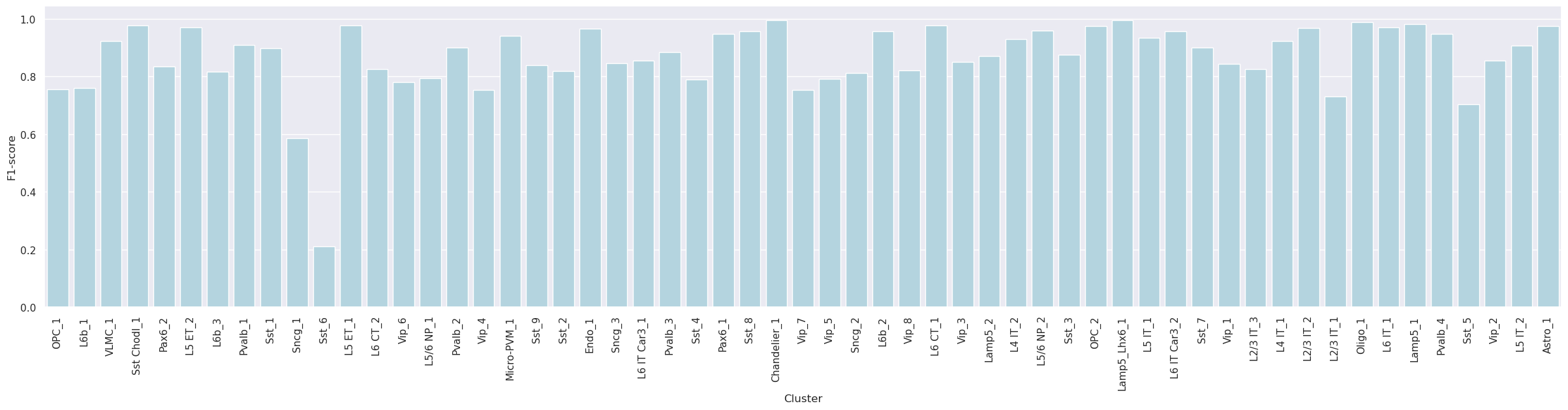
-
Confidence values for correctly and incorrectly assigned labels
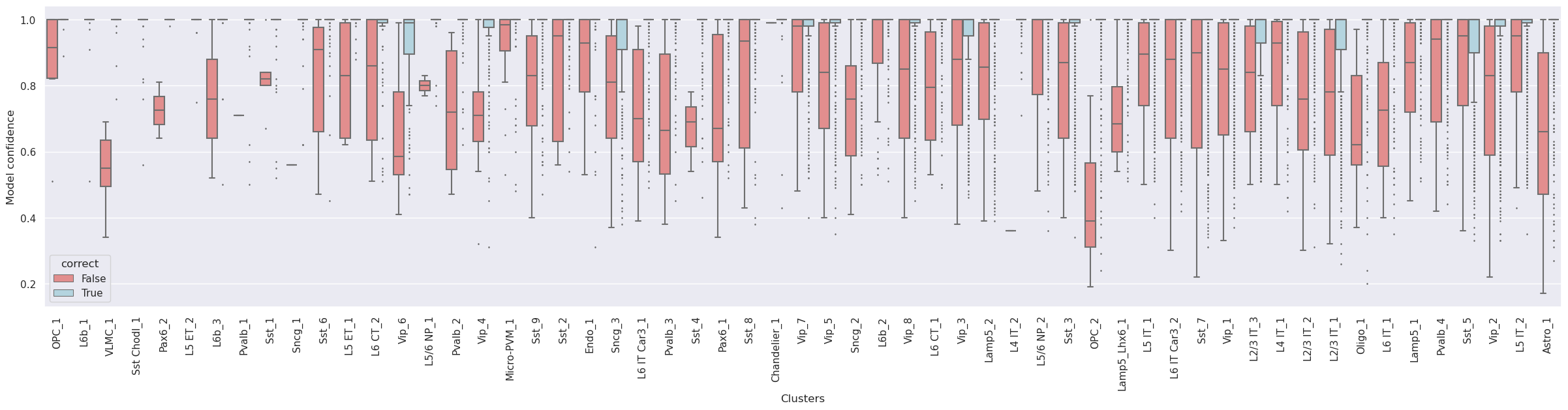
-
Confusion matrix (row-normalized)